Version 4.9.5 released (February 2019)
- Haemopedia RNA-seq now published at Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans NAR (2019)
- RNA-seq data is now normalised as TPM (transcripts per million) rather than RPKM (reads per kilobase million)
- A tour of Haemosphere can be accessed by clicking the question marks
- Gene expression can now be viewed across multiple datasets and species.
- Gene vs gene expression plots are now available.
Haemosphere is a web portal to explore gene expression data on haematopoietic cell types.
Features include differential expression analysis, gene set management and visualisation tools.
more...

Search genes based on symbol or synonym matches or by uploading a list of genes.
Perform differential expression analysis using R/Bioconductor as analysis engine.

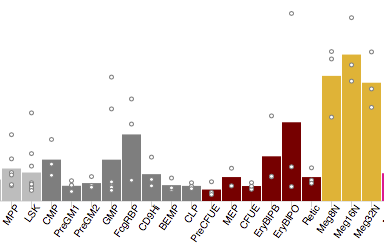
Haemosphere acts as a single point of entry for some key datasets of value to haematopoiesis
research community. View datasets and perform principal components analysis on cell types.


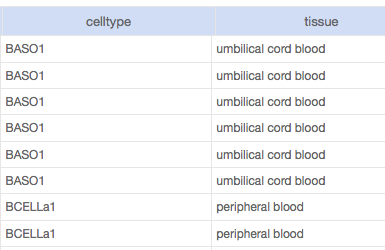
View metadata for each dataset, such as cell type, tissue of origin, mouse strains or FACS markers.